This is an old revision of the document!
Table of Contents
What is NGS?
Technological advances in molecular genetics at the end of the twentieth century have established a strong foundation for genetic analysis. Shotgun sequencing has allowed for the sequencing of longer DNA sections, which has played a significant role in the Human Genome Project (cite). In this method, DNA is enzymatically broken into smaller fragments and cloned for individual sequencing prior to realignment (cite). Nearly a decade later, subsequent advances in next generation sequencing (NGS) technologies have made genomic analysis much more economically feasible and have thus enabled applications of genomics across clinical and research settings (cite).
Different Types of NGS Technology
454 Life Sciences
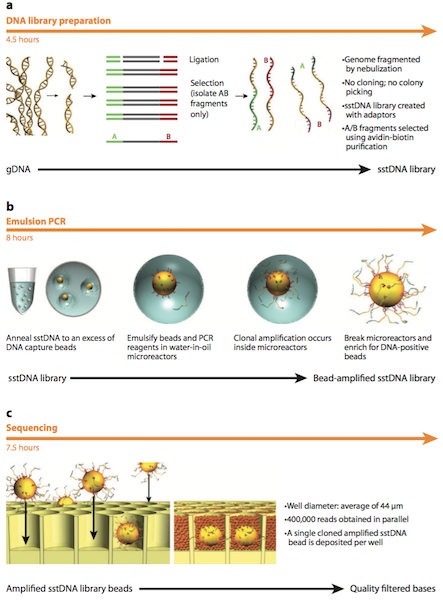
In 2005, the first Next Generation Sequencing (NGS) system became available on the market by 454 Life Sciences. As described by Margulies et al. (2005), this technique first includes fragmentation of the genome and addition of adapters to the fragments to create an adapter ligated single stranded DNA library.3 The DNA library is mixed with capture beads which are in an amplification microreactor. PCR reagents and emulsion oil is then added in preparation for emulsion PCR. Once PCR has been conducted, the microreactors are broken and the beads containing DNA are isolated. The DNA beads are then deposited into a PicoTiter Plate with enzyme beads and are subjected to centrifugation. Next, simultaneous sequencing of the entire genome occurs in the wells through a pyrophosphate-based sequencing method. This sequencing method detects the release of an inorganic pyrophosphate (PPi) molecule when a nucleotide is incorporated. Sulfurylase then converts adenosine 5’-phosphosulfate (APS) and PPi into adenosine triphosphate (ATP). ATP and luciferin are subsequently used by luciferase to convert luciferin to oxylucifern and produce light. The photons are captured by a charge-coupled device (CCD) and quantified to determine the sequence of the fragment. More specifically, a base calling method is used to determine the DNA sequence from the signals that are captured. Each well has template-carrying beads which have a known four-nucleotide sequence. This sequence, which is present at the beginning of the flowgram, is used to determine the DNA sequence. The intensity of each peak in the flowgram is proportional to the number of nucleotides in the sequence.3
Applied Biosystems SOLiD
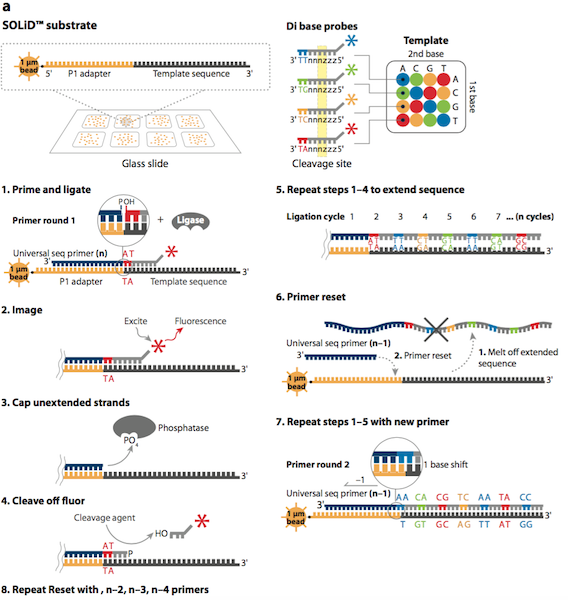
Sequencing by Olignucleotide Ligation and Detection (SOLiD) is a next generation sequencing technique developed by Life Technologies and purchased by Applied Biosystems in 2006.4 SOLiD was made available for commercial use in the same year. This method uses a two-base fluorescent probe, which is based on sequencing by ligation. Sequencing by ligation uses DNA ligase to identify the nucleotide present at a given position in a DNA sequence.4 Ligation sequencing does not use a DNA polymerase to generate a second complimentary strand, but uses mismatch sensitivity of ligase to determine the sequence of DNA.4
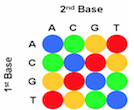
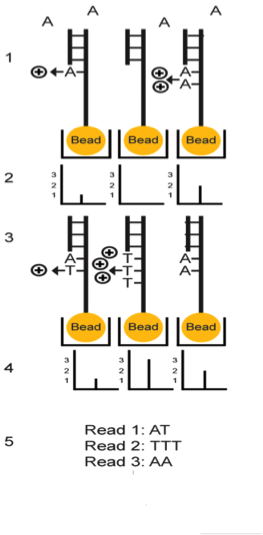
The first step in the SOLiD sequencing system is library preparation. The are two common methods of preparing a DNA library. The first involves fragmentation while the other uses a mate-paired library. In the first process, the whole genome of the target DNA is randomly fragmented and two different 25 base pairs (bp) DNA adaptors on beads are ligated at the 5’ and 3’ ends.5 Mate-paired library DNA fragments, however, are separated from another DNA fragments of known length using an internal adaptor. This process also includes DNA adaptors at the 5’ and 3’ end similar to the fragmentation method.5 Next, DNA fragments on the beads are amplified by the emulsion polymerase chain reaction (PCR) technique. The beads containing clonally amplified complementary DNA fragments attach to the surface of a flow cell.5 Sequencing of the target DNA sequence involves multiple rounds of ligation reactions. In the first round, a universal sequencing primer is annealed to the adaptor and 16 fluorescently labeled octamer oligonucleotides are added.6 The 16 possible oligonucleotides represent all the possible combinations of two nucleotides such as AT, AG, AC and so on in the first two bases while the 3rd to 5th bases are degenerate and unknown.6 The last 3 bases are bound to one of the four fluorescent labels, which are different in the emission spectrum.6 Each label represents four dinucleotides in each ligation reaction and only the first two nucleotides in the probe are used to help identify the sequence of the DNA fragment. Since each label represents four possible dinucleotide combinations, the first round of annealing a probe to the primer does not exactly identify the bases specifically, but allows the possible sequence to be narrowed down.6,7 The final three bases are then cleaved off after detection of the fluorescent label in order to provide a free 5’ end for the ligation of the next probe. This ligation process continues for approximately 5-7 cycles.7 After the cycles of ligation, fluorophore detection, and cleavage, the previous ligated probes are denatured and removed. A new primer that is shorter by one base (n-1) is annealed to the template and another 5-7 cycles of ligation, detection and cleavage occurs. This allows the detection of up to 75 base pairs depending on the specific SOLiD sequencing software used after multiple rounds have been completed.5
Multiple cycles and rounds allows for the interrogation of every base in two different ligation reactions in addition to two different primers. This process is called double interrogation and it is effective at increasing accuracy and specificity of the template strand.7,8 The fluorescent detection and visualization is analyzed using a colour space reference sequence as opposed to a nucleotide based reference sequence.7,8 In order to analyze the data from the detected fluorescence, extended algorithm software programs based on colour space are used. There are multiple programs that are available in the market for analyzing SOLiD data.8
Figure 4 - A summary of SOLiD sequencing steps are illustrated in this video.
Illumina Sequencing Technology
Illumina sequencing technology is one of the most successful and widely adopted next-generation sequencing (NGS) technologies worldwide. It enables a wide variety of applications, allowing researchers to ask virtually any question related to the genome, transcriptome (the full range of messenger RNA molecules expressed by an organism), or epigenome (several chemical compounds that can tell the genome what to do).9,10
Sample Prep
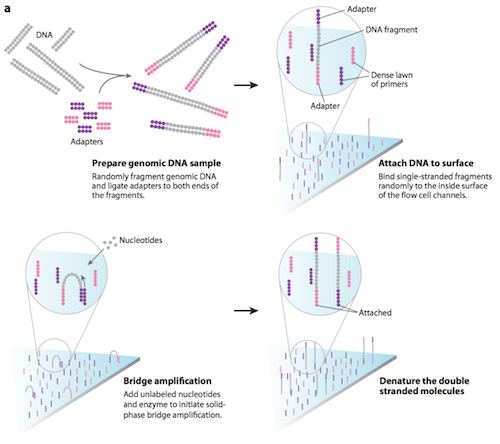
The process of sample preparation begins with tagmentation. Transposons randomly break up the genomic DNA into more manageable fragments of around 200 to 600 base pairs, then tag both ends of the DNA fragments with adaptors (a short, chemically synthesized, double-stranded DNA molecule which is used to link together two other DNA molecules). Following tagmentation, the DNA fragments attached to the adaptors are made into single strands.11 Once prepared, the DNA fragments are washed across the flow cell (a glass slide with lanes). The complementary adaptor regions on DNA fragments bind to primers (short strand of RNA that acts as a starting point for DNA replication) on the surface of the flow cell. DNA fragments that do not attach, are washed away.11
Cluster Generation
During cluster generation, the DNA attached to the flow cell is replicated to form small clusters of DNA with the same sequence. The process begins with unlabelled nucleotide bases and DNA polymerase being added, to lengthen and join the strands of DNA attached to the flow cell. This initiates solid-phase bridge amplification. 11 The polymerase incorporates nucleotides creating ‘bridges’ of double-stranded DNA between the primers on the flow cell surface. The double-stranded DNA is then denatured leaving single-stranded templates anchored to the flow cell. The process is repeated over and over generating several million dense clusters of identical DNA sequences.11
Several million clusters can be amplified to distinguishable locations within each of eight independent 'lanes' that are on a single flow-cell. This means that eight independent libraries can be sequenced in parallel during the same instrument run.12
Sequencing
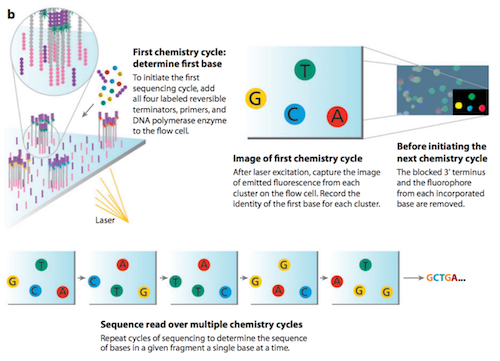
The method of sequencing utilized by Illumina is called sequencing by synthesis.11 The first sequencing cycle begins by adding four fluorescently-labelled reversible terminator nucleotides, primers, and DNA polymerase to the flow cell. The primer attaches to the DNA being sequenced. The DNA polymerase then binds to the primer and adds the first fluorescently-labelled terminator nucleotide (base) to the new DNA strand. Terminator nucleotides have been modified in two ways. First, they are “reversible terminators”, in that, a chemically cleavable moiety at the 3’ hydroxyl position allows only a single-base incorporation to occur in each cycle. Second, one of four fluorescent labels, also chemically cleavable, corresponds to the identity of each nucleotide (A, C, G, or T).12 Once a terminator base has been added, no more bases can be added to the strand of DNA until the terminator moiety is cut from the DNA. Lasers are passed over the flow cell to activate the fluorescent label on the nucleotide base. This fluorescence is detected by a camera and recorded on a computer. Each of the terminator bases (A, C, G and T) emit a different colour. The emitted fluorescence from each cluster is captured and the first base is identified.The fluorescently-labelled terminator group is then removed from the first base. This allows for the next sequencing cycle to begin with the next fluorescently-labelled terminator base being added alongside the first base. After laser excitation, the image is captured as before, and the identity of the second base is recorded. The sequencing cycles are repeated to determine the sequence of bases in a fragment, one base at a time. The process continues until millions of clusters have been sequenced.11
Data Analysis
The sequence generated is then aligned with a reference sequence. This looks for matches or changes in the sequenced DNA.11
Ion Torrent Sequencing
Like other next generation sequencing techniques, Ion Torrent also requires sequence amplification, but it is the first technique that uses electrochemical detection, not camera scanning and fluorescence detection. Ion Torrent is done with an Ion Personal Genome Machine (PGM), which was released near the end of 2010. With quick turnover rates that have limited quantitative data and its small instrument size, Ion Torrent has quite an efficient cost due to the PGM.4
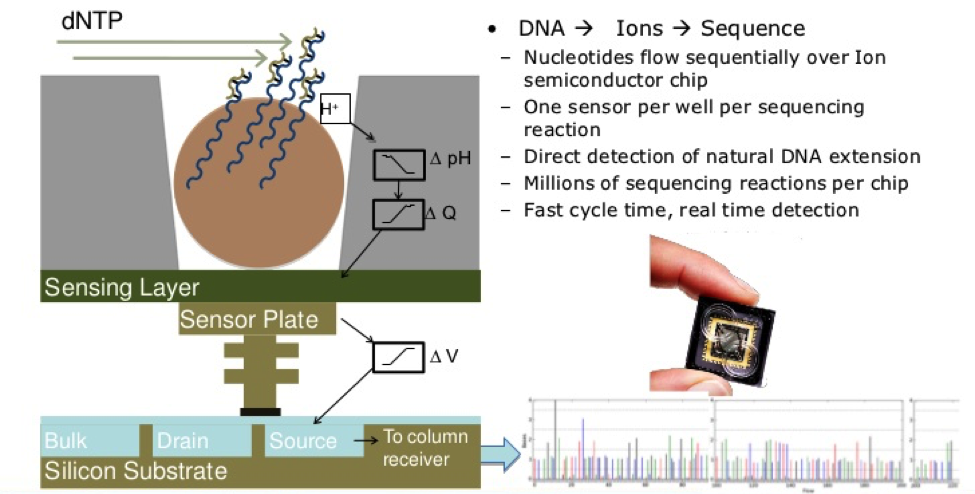
Firstly, the DNA sample is prepared from fragmented RNA and then linked together.13 The library is then clonally amplified by emulsion PCR onto beads known as Ion Sphere Particles. The beads are placed into proton-sensing wells on a semiconductor sequencing chip so that one bead can roughly fit into hundreds of millions of wells.14 The chip is then submerged into a nucleotide solution and results in the release of protons and a corresponding change in pH. This change in pH is documented by the PGM to determine whether the right nucleotide was used in the process or not, let alone if a nucleotide was added at all.13 As sequencing occurs, each of the four bases is introduced sequentially.14 A clear indication of the correct nucleotide being added is the presence of voltage. There will be no voltage found if the wrong nucleotide is added, and there will be double the voltage if two nucleotides are added.4 The more nucleotides present, the greater the increase in voltage and pH.15 It is important to note that Ion Torrent sequencing is very fast and allows for many more reads to be done per sequencing run as there is no optical detection necessary.13
However, there can be error that results from Ion Torrent sequencing. When trying to distinguish between 7-8 nucleotides, it becomes a bit more challenging which is why error rates can go as high as 1.7%.13 The reason for this is because of the homopolymer sequences (a sequence of identical bases) being used. As the homopolymer length increases, the deletion error rate increases and the insertion error rate stays relatively constant. This causes in a large release in protons, and when a certain concentration is exceeded the pH readings start to become ambiguous which causes insertion or deletion. These errors could be much larger if not for the Ion Torrent’s short read lengths which mitigates the number of successive nucleotides being read by these errors.15
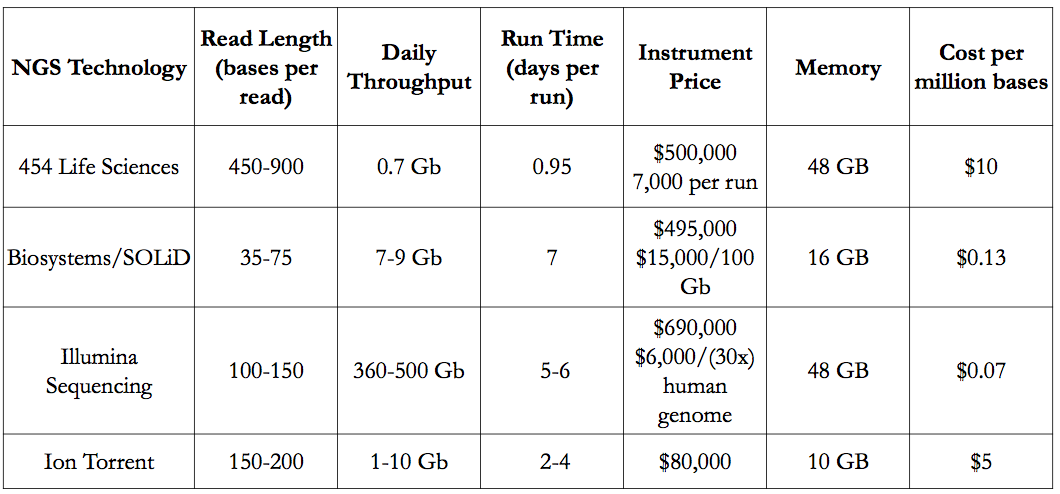
Advantages and Disadvantages of The 4 Types of NGS
Applications of NGS
Oncology
Next generation sequencing has lead to significant advances in cancer diagnostics as tumour subtypes can now be identified through genetic testing instead of morphological characteristics.16 NGS is also used in finding appropriate cancer therapies as well as identifying resistance mutations when patients stop responding to cancer treatments.16 More specifically, NGS technology has been applied to the detection of lung adenocarcinomas through identifying genomic alterations in tumours.17 Recently, NGS methods have also been used in brain tumour diagnostics. Sahm and colleagues (2015) report on the use of NGS to detect mutant alleles in neuropathology samples.18 NGS technology used to detect single nucleotide variations in genes associated with specific cancers is especially important in personalized medicine and therapies. For example, for certain clinical trials, the tumour genetic makeup needs to be well established before an individual can participate in the trial, therefore, NGS is used in the genetic profiling of the tumour.16These patients can then be directed to the appropriate clinical trial or they can have therapies personalized to target their specific cancer.16
Ecology and Biodiversity
NGS technologies have numerous applications outside of human genetic testing. In ecology, the mass sequencing of environmental samples remain a priority in biodiversity research.19 With NGS, it has facilitated the analysis of environmental samples derived from a variety of ecosystems. Most of the ecological studies and research aim to discover the species or biological material present in a certain environment. NGS technology increases the capacity at which massive amounts of DNA data is produced at an improved efficiency and specificity compared with traditional Sanger sequencing methods.19 Most of the studies that use NGS technology in their research employ the 454 pyrosequencing platform mainly because it has a longer sequence read length.19 In addition to examining the biodiversity of DNA samples, ecologists can also observe the slight changes in community structure that may occur during anthropogenic or natural environmental fluctuations. The data retrieved from NGS can describe the ecosystem health and stability. For example, several studies were conducted using NGS technology on old terrestrial environmental samples in order to provide more evidence on evolutionary ecology. Overall, NGS can identify species in a variety of ecosystems that range from freshwater, marine, soil terrestrial and gut microbiota.19
Ion Torrent PGM - Microbial Pathogens
One of the more interesting applications of Ion Torrent PGM sequencer is how it can be used to identify microbial pathogens. In May and June of 2011, a toxin known as Shiga-toxin, which produces Escherichia coli (E. coli) was having an ongoing outbreak in Germany. More than 3000 people were infected. With the recent advancements in NGS systems this specific incident showcased the efficiency and progress that Ion Torrent PGM sequencer has made. The whole genome sequencing of the Ion Torrent PGM sequencer allowed for the scientists to find the specific antibiotic resistances needed to by identifying which type of E. coli was present. What was revealed was the presence of a hybrid of 2 E. coli strains. One being entero aggregative E. coli and the other being entero hemorrhagic E. coli. The presence of a hybrid of 2 E. coli strains seemed to help explain why the outbreak had behaved in a pathogenic manner. Based on the sequencing results found from the E. coli, Ion Torrent PGM provides a fast method of sequencing for when an outbreak of a new disease occurs.4
Direct to Consumer Genetic Testing
Advancements in terms of the scale and efficiency of genetic sequencing has decreased the cost of using next generation technologies. This decrease in cost has allowed the power of these high-throughput DNA sequencing technologies to be harnessed by companies, like 23andMe, which offer direct-to-consumer (DTC) genetic testing.20
23andMe is a company that uses the Illumina HumanOmniExpress-24 format chip (a form of Illumina next generation sequencing), to sequence DNA, and subsequently analyze it to provide genetic profiling information directly to consumers.21 One simply has to pay $199, and provide a saliva sample. The information provided is divided into four categories: carrier status, wellness, traits, and ancestry. Carrier status indicates whether a person carries genes associated with any of 36 different disorders, such as, cystic fibrosis. Wellness addresses aspects including, whether an individual is lactose intolerant, or if they tend to get red and flushed when they drink alcohol. The traits information looks at a set of nearly two dozen traits that are dictated by genes including, male-patterned baldness, eye colour, and hair colour. Finally, ancestry information is also provided, including a breakdown of the percentage, by genetic makeup, of different racial and ethnic groups in an individual’s background. Finally, ancestry information is also provided in regards to, ancestry composition (a breakdown of the percentage, by genetic makeup, of different racial and ethnic groups in an individual’s background), DNA relatives, maternal and paternal lineages, and even neanderthal percentage.21
However, services such as those offered by 23andMe are not left unregulated. Until November 2013, 23andMe offered consumers genetic testing to estimate their risk for 240 heath conditions, such as breast cancer and heart disease, for $99.22 In November 2013, the FDA shut down the company after it was determined that the company lacked the appropriate approval to give people potentially life-altering information about their health. Of particular concern were assessments such as those for BRCA-related genetic risk and drug responses (e.g., warfarin sensitivity) because of the potential health consequences that could result from false positive or false negative assessments for these high-risk indications.23 For instance, “if the BRCA-related risk assessment for breast or ovarian cancer reports a false positive, it could lead a patient to undergo prophylactic surgery, chemoprevention, intensive screening, or other morbidity-inducing actions, while a false negative could result in a failure to recognize an actual risk that may exist”.23 The FDA’s other concern related to how people were interpreting the reported results. Some people found the information confusing; misinterpreting the presence of a genetic variant that is simply linked to a disease, with a definite expectation of developing the disorder.23 After implementing changes to address the FDA's concerns, nearly 2 years later in October 2015, 23andMe released the current FDA approved, previously mentioned, $199 test (a scaled back version of the original).24
The applications of next-generation sequencing seem almost endless, allowing for rapid advances in many fields including the field of consumer genetic testing.
DID YOU KNOW?
Breakthrough Technology: In 2014, Illumina announced the introduction of HiSeq X, a new high-throughput sequencer that will allow for whole genome sequencing for $1000. The applications of the HiSeq X sequencer, however, remains for research purposes and the $1000 whole genome sequencing cost has yet to be incorporated in clinical laboratories (Hayden, 2014).
References
1.
2.
3. Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., … Rothberg, J. M. (2005). Genome Sequencing in Open Microfabricated High Density Picoliter Reactors. Nature, 437(7057), 376–380.
4. Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., … & Law, M. (2012). Comparison of next-generation sequencing systems. BioMed Research International, 2012.
5. Ansorge, W. J. (2009). Next-generation DNA sequencing techniques. New Biotechnology, 25(4), 195-203.
6. Corney, D. (2013). RNA-seq Using Next Generation Sequencing. Materials And Methods, 3, 203. http://dx.doi.org/10.13070/mm.en.3.203
7.Metzker, M. L. (2010). Sequencing technologies—the next generation.Nature reviews genetics, 11(1), 31-46.
8. Breu, H. (2010). A theoretical understanding of 2 base color codes and its application to annotation, error detection, and error correction. White paper, Life Technologies.
9. NIH. (2015). Epigenomics Fact Sheet. Retrieved January 21, 2016, from https://www.genome.gov/27532724
10. Nature. (n.d.). Transcriptome. Retrieved January 21, 2016, from http://www.nature.com/scitable/definition/transcriptome-296
11. Illumina. (2010). Illumina Sequencing Technology. Retrieved January 21, 2016, from https://www.illumina.com/documents/products/techspotlights/techspotlight_sequencing.pdf
12. Shendure, J., & Ji, H. (2008). Next-generation DNA sequencing. Nature Biotechnology, 26, 1135-1145. Retrieved January 21, 2016.
13. Corney, D. C. (2013). RNA-seq Using Next Generation Sequencing. Retrieved January 17, 2016, from http://www.labome.com/method/RNA-seq-Using-Next-Generation-Sequencing.html
14. Quail, M. A., Smith, M., Coupland, P., Otto, T. D., Harris, S. R., Connor, T. R., … & Gu, Y. (2012). A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics, 13(1), 341.
15. Kremkow, B. G., & Lee, K. H. (2015). Sequencing technologies for animal cell culture research. Biotechnology Letters, 37(1), 55-65.
16. Gagan, J., & Van Allen, E. M. (2015). Next-generation sequencing to guide cancer therapy. Genome Medicine, 7(1), 1-10.
17. Drilon, A., Wang, L., Arcila, M. E., Balasubramanian, S., Greenbowe, J. R., Ross, J. S., … & Ladanyi, M. (2015). Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in“ driver-negative” lung adenocarcinomas. Clinical Cancer Research, clincanres - 2683.
18. Sahm, F., Schrimpf, D., Jones, D. T., Meyer, J., Kratz, A., Reuss, D., … & Buchhalter, I. (2015). Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta neuropathologica, 1-8.
19. Shokralla, S., Spall, J. L., Gibson, J. F., & Hajibabaei, M. (2012). Next‐generation sequencing technologies for environmental DNA research. Molecular Ecology, 21(8), 1794-1805.
20. Nature. (2015). Applications of next-generation sequencing. Retrieved January 21, 2016, from http://www.nature.com/nrg/series/nextgeneration/index.html
21. 23andMe. (2015). 23andMe Canada - DNA Genetic Testing & Analysis. Retrieved January 21, 2016, from https://www.23andme.com/en-ca/
22. Hayden, E. (2015). Out of regulatory limbo, 23andMe resumes some health tests and hopes to offer more. Retrieved January 21, 2016, from http://www.nature.com/news/out-of-regulatory-limbo-23andme-resumes-some-health-tests-and-hopes-to-offer-more-1.18641
23. FDA. (2013). 23andMe, Inc. 11/22/13. Retrieved January 21, 2016, from http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/2013/ucm376296.htm
24. Park, A. (2015). Genetic Testing Company 23andMe Returns to Market. Retrieved January 21, 2016, from http://time.com/4080583/23andme-dna-genetic-testing/
25. Hayden, E. C. (2014). The $1,000 genome. Nature, 507(7492), 294-295.
Images retrieved from:
Mardis, E. R. (2008). Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet., 9, 387-402.
Tinning, M. (2012, August 1). NGS technologies - platforms and applications. Retrieved January 20, 2016, from http://www.slideshare.net/AGRF_Ltd/ngs-technologies-platforms-and-applications