Table of Contents
DAROBACTIN
The paper titled "A new antibiotic selectively kills Gram-negative pathogens" can be found by the link.
Introduction
Infectious diseases are a significant cause of death worldwide, accounting for approximately one third of all deaths among the top ten leading causes of death (Center for Disease Control (CDC), 1999). Currently, the main treatment for infectious diseases is prescribing antibiotics; however, the rate of resistance to antibiotics by bacteria is increasing whilst the production of new antibiotics is slowing down (Lewis, 2013). Bacteria can mutate and evolve which, over time, allows them to develop resistance to currently prescribed antibiotics. This is especially an issue for Gram-negative bacteria as they have an outer membrane that acts as a protective layer which limits the transfer of compounds.
Researchers theorized that useful compounds can be found in microorganisms that have the same requirements for antibiotics as humans. They identified Photorhabdus and Xenorhabdus as potential bacteria as these microorganisms produce antimicrobials to compete with Gram-negative bacteria in their environment. Imai et al. set out to identify the compound with antimicrobial properties, which they named darobactin, as well as determine its target. The investigators would also measure the efficacy of darobactin in animal models.
Methods
Identifying darobactin
This investigation began with the identification of darobactin by screening a small set of Photorhabdus and Xenorhabdus strains against E. coli. On the strain that produced a zone of inhibition, high performance liquid chromatography was used to extract an active fraction of darobactin. High-resolution electrospray ionization–Mass spectroscopy was used to determine the exact mass of darobactin, while NMR studies were used to determine the structure of the active compound. They created four possible darobactin isomers that underwent conformational stretching. The lowest energy isomer for each conformation was then subjected to geometry optimization. After determining the structure of darobactin, it was necessary to identify the biosynthetic gene cluster (BGC) encoding this peptide. To identify the BGC, the genome of P.khanii HGB1456 was sequenced. The results were then analyzed using antibiotic and secondary metabolite analysis shell. BLAST was then used to screen for the core peptide sequence WNWSKSF on all Photohabdus genomes. To identify other bacterial species that may produce darobactin-like compounds, homologous enzymes were searched using the DarE sequence in BLAST
Identifying darobactin target
The researchers tested the activity of darobactin against a range of Gram-negative bacteria by determining minimum inhibitory concentrations (MIC). Overnight cultures of various strains were created, diluted and subsequently added to darobactin solutions. After overnight incubation, the minimum inhibitory concentration was found to be the concentration where no growth of strains can be detected by the eye. In addition, the cytotoxicity of darobactin was determined through a microplate Alamar blue assay. Next, to identify the target of darobactin, mutants that were resistant to darobactin were obtained. Darobactin resistant mutants were generated by the daily serial passage of E. coli at sub-MIC concentrations, leading to a steady shift in the darobactin concentration that allows E. coli growth. To observe the direct inhibition of the darobactin target, a protein refolding assay was used in which the function of the target was measured. Once the particular target of darobactin was found, the researchers used high resolution NMR to characterize the interaction of BamA at the atomic level.
Using animal models
Due to the promising results of darobactin, the researchers tested the efficacy in mouse models. Female CD-1 mice were used in a non-randomized nor blinded experiment. Four different models were used. In the virulence model both wild type and BamA mutations leading to darobactin resistance were tested in an acute infection model. 24 hours after the infection, the mice were euthanized and the spleen and liver were analyzed. To understand the pharmacokinetics, mice were injected with darobactin intraperitoneally and blood samples were collected at different time points. In the septicaemia model, mice were tested for protection against both wild type and multidrug resistant E.coli, as well as other strains of bacteria. While test mice would receive a dose of darobactin after the infection, control mice received a dose of gentamicin. In the thigh infection model, darobactin was tested in a neutropenic thigh infection model against multidrug-resistant E. coli. Cyclophosphamide was utilized to render mice neutropenic. After infection, darobactin was administered and the right quadriceps were analyzed. To determine the significance of their results, the authors carried out statistical analyses and determined the confidence intervals.
Structure
Darobactin is a large molecule (molecular mass of 965 Da) classified as a modified heptapeptide with a linear sequence that is twice cyclized (Trp1-Asn2-Trp3-Ser4-Lys5-Ser6-Phe7) (Figure X.) (Imai et al., 2019; Kaur et al., 2021).
 Figure 1. Structure of darobactin as determined by Imai et al. (2019).
Figure 1. Structure of darobactin as determined by Imai et al. (2019).
Through NMR studies, Imai et al. (2019) were able to determine the structure of darobactin, specifically they identified two uncommon macrocycle crosslinks. The first is between the C7 indole of Trp1 and the β-carbon of Trp3 which forms an aromatic-aliphatic ether link. The second crosslink is a bond between the C6 indole of Trp3 and the β-carbon of Lys5 - this bond is novel for antibiotics because it is between two un-activated carbons (Imai et al., 2019). These un-activated carbons are linked via a radical S-adenosylmethionine (SAM) enzyme which is encoded by the dar operon. These crosslinks create two chiral centers at Trp3 (R configuration) and Lys5 (S configuration) (Imai et al., 2019).
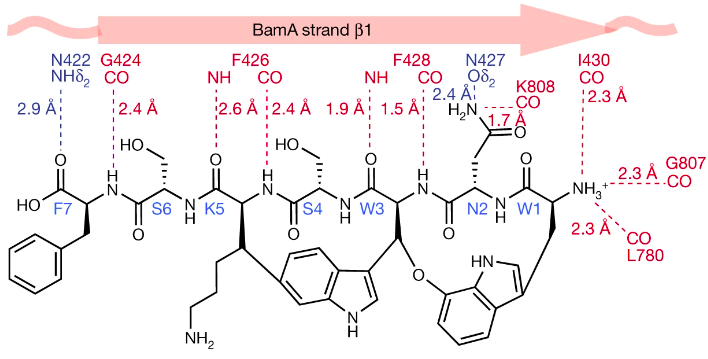
Furthermore, Kaur et al. (2021) investigated the structure of darobactin in relation to its binding with BamA. In particular, they explored the importance of the backbone contact between darobactin and BamA. It was found that darobactin forces BamA into an open state by binding to its lateral gate via hydrogen bonds in an antiparallel beta sheet conformation (Kaur et al., 2021). Hydrogen bonds ranging from Ile430 in BamA and Trp1 in darobactin, continue to Gly424 and Phe7 (Kaur et al. 2021). Additional hydrogen bonds are formed between the N-terminus of darobactin and Ile430 in strand β1, Gly807 in β16 and Leu780. Using this information, Kaur et al. (2021) determined that once bound to BamA, darobactinA mimics the beta-strand that opens the BamA lateral gate. Hence, the structure of darobactin, specifically its backbone that makes contact with BamA, is highly important in its antibiotic function.
In addition, darobactin’s structure plays an important role in its efficacy against resistance mutations. Through mutant studies, Kaur et al. (2021) found that the amino acid composition of darobactin is variable and its binding site sequence is not conserved in homologues of BamA, making it effective against mutations. The backbone contacts that dominate the binding between darobactin and BamA suggest that changes to the BamA sequence will have minimal effects on darobactin (Kaur et al. 2021). Kaur et al. produced alanine mutants of BamA, and tested the affinity of darobactin against the mutant. None of the side chain contacts were conserved between darobactin A and the mutated BamA (Kaur et al. 2021). Despite the changes, the kD of the interaction between darobactin and the mutant was nearly identical to that of the darobactin and wild-type BamA (Kaur et al. 2021). Moreover, the composition of darobactin’s binding pocket is unusual as it contains peptidic, lipidic as well as aqueous components (Kaur et al., 2021). This indicates that darobactin has evolved to fit into the peptido-lipidic environment of the BamA lateral gate (Kaur et al. 2021).
Mechanism
Utilizing cryo-electron microscopy reconstruction at 3.0 Å resolution, researchers were able to determine the positional attachments of darobactin onto BamA and the subsequent BamA positional changes revealing the positional mechanisms of how darobactin is able to interfere with BamA function. Upon visualization, researchers found that darobactin binds to BamA lateral gate in antiparallel beta-sheet conformation to the B1 strand with a series of backbone hydrogen bonds. The binding pocket of darobactin suggests that it acts as a molecular mimic of a β strand that seals the open lateral gate of BamA upon binding. The observations that darobactin seals the gate open state of BamA is additionally confirmed by comparison of 2D transverse relaxation optimized spectroscopy, also known as TROSY, NMR spectra. The spectroscopy compared the fingerprint spectra of different BamA preparations in the absence and presence of darobactin. These findings were comparable to that of the native closed-gate conformation of BamA when stabilized by darobactin. This suggests that darobactin stabilization is able to confer a conformational shift closing the lateral gate preventing the exit of substrates into the outer membrane by blocking the first step of the insertion reaction.
In its native function, BamA is able to recognize the consensus β signal located in the other membrane nascent chain and anchor its conserved terminal aromatic residue to the lateral gate. Once anchored, BamA would be able to fold and assemble the nascent chain and insert it into the outer membrane. However, darobactin binds specifically to the dynamic lateral gate of BamA, blocking the β signal binding site for the outer membrane protein nascent chains. Most BAM substrates contain the same conserved aromatic residue in its consensus signal sequence at the C terminus which have evolved to interact only transiently with the gate region of BamA. Darobactin is able to outcompete all substrates with higher binding affinity, preventing recognition from the consensus β-signals in the novel nascent chains. By binding onto the β signal binding site and inhibiting the insertion reaction catalyzed by BamA, darobactin is able to target the bacterial membrane's process of bacterial outer membrane protein biogenesis.Without BamA, the nascent chains are unable to fold and assemble correctly nor able to exit into the outer membrane (Figure 3). Loss of its outer membrane proteins, gram-negative bacteria are unable to protect itself against the harsh environment as well as crucial processes such as translocation, signal transduction, and overall bacterial membrane stability. (Imani et al, 2019; Kaur et al., 2021).
Results
In the study conducted by Imai et al. (2019), results were collected based on identifying the unknown at the time drug darobactin and the identification of the target of darobactin in gram-negative bacteria.
Darobactin Identification
The authors first looked at strains of Photorhabdus and Xernorhabdus, screened against E. coli (Imai et al., 2019). Due to the existence of ‘silent’ biosynthetic gene clusters (BGCs) that were not expressed very well in vitro, few inhibition zones were observed, so concentrated extracts from the strains were instead used (Imai et al., 2019). Doing so, they found that the extract of Photorhabdus khanii HGB1456 formed inhibition zones (Imai et al., 2019).
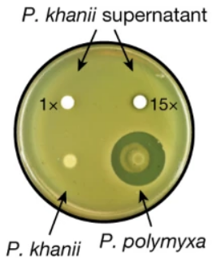
Figure 4: Inhibitory ability is observed in the concentrated extract of P. khanii when plated with E. coli MG1655, forming inhibition zones, whereas the strain itself does not produce these (Imai et al., 2019). P. polymyxa is used as the positive control for comparison (Imai et al., 2019).
Using this, the authors looked into what was causing this inhibitory activity. They began by using HPLC to isolate the active extract and used electrospray ionization mass-spectroscopy to determine that the causative compound was one with a molecular mass of 966.41047 g/mol (Imai et al., 2019).
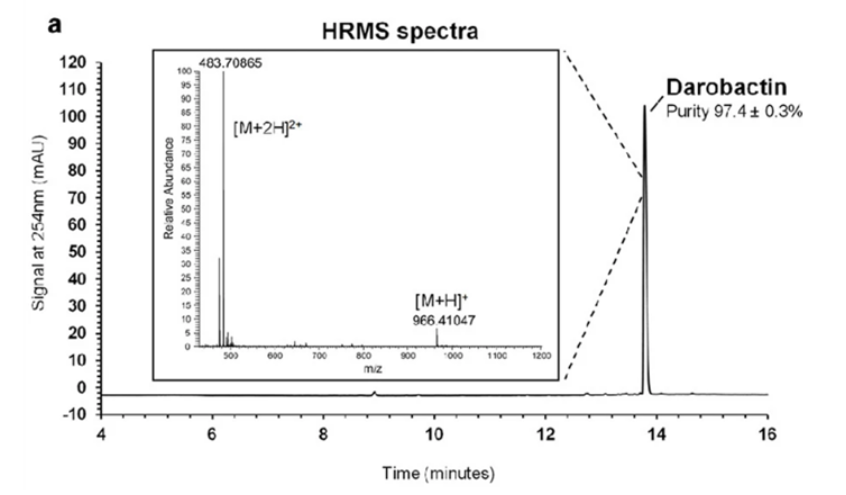
Figure 5: The high-resolution mass spectrum and HPLC chromatogram produced aided in the determination of darobactin (Imai et al., 2019). HPLC allowed for darobactin to be separated from the matrix with a purity of 97.4 ± 0.3% and HRMS gave the total molar mass of darobactin (Imai et al., 2019).
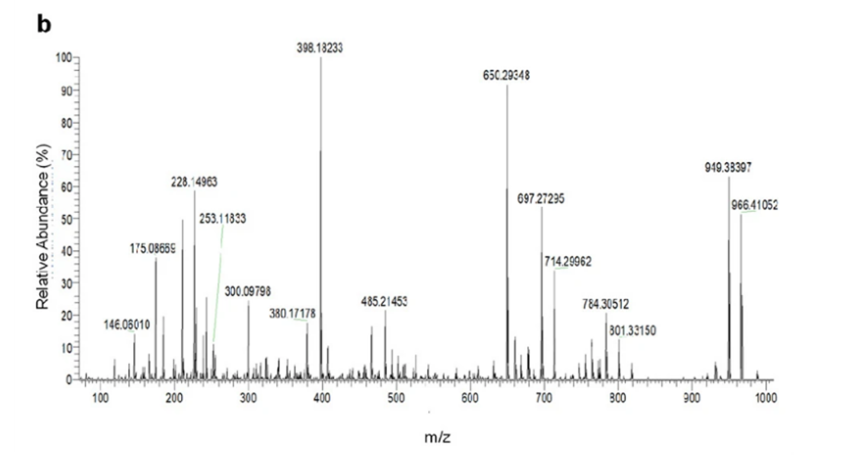
Figure 6: MS/MS performed gave various masses corresponding to fragments formed in the process. Analysis of the mass spectrum fragments alongside information retrieved from NMR, the structure and formula of darobactin were determined (Imai et al., 2019).
The authors derived the chemical formula as C47H56O12N11+ and were unable to find matches in Antibase based on the mass of this product (Imai et al., 2019). Concluding that this must be some novel product, the authors used NMR and mass-spectroscopy to determine the structure, calling it darobactin (Imai et al., 2019).
Target Identification
Darobactin was found to act well against various gram-negative bacteria having an MIC of 2 ug/mL against some drug-resistant E. coli and K. pneumoniae (Imai et al., 2019). In fact, the minimum bactericidal concentration (MBC) was found to be 8 ug/mL with respect to E. coli (Imai et al., 2019). Compared to ampicillin, darobactin’s MBC is at least 64 times less in E. coli which is indicative of its efficacy in fighting against gram-negative bacteria (Imai et al., 2019). However, darobactin did not appear to act as well when used on gram-positive or gut bacteria (Imai et al., 2019).
Due to its size of 965 Da being greater than the 600 Da size limit for entry through the outer membrane of gram-negative bacteria, darobactin is unable to enter the outer membrane (Imai et al., 2019). The authors inferred that this allowed darobactin to act on these bacteria like polymyxin through targeting of lipopolysaccharides (LPS) and tested to see if this was the case (Imai et al., 2019). To do so, LPS was added to samples of bacteria treated with polymyxin or darobactin. This resulted in the effects of polymyxin being rescued, as expected, while darobactin treated bacteria were still killed, implying a different mechanism being used (Imai et al., 2019). Notably, there was welling in the membrane of cells treated with darobactin which led to their lysis (Imai et al., 2019).
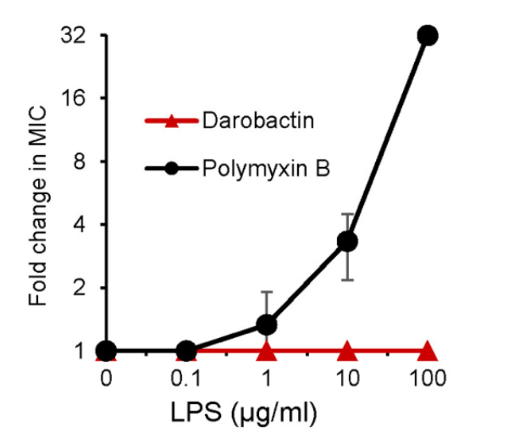
Figure 7: Addition of LPS resulted in an increased resistance to Polymyxin B, however, no such change in resistance was observed corresponding to darobactin (Imai et al., 2019).
Transcriptome analysis and ligand protection thermal proteome analysis were performed in hopes of discovering a mechanism in which darobactin targets (Imai et al., 2019). Results from transcriptome analysis showed the induction of sigma E and Rcs envelope stress responses however, ligand protection thermal proteome analysis was unable to determine a specific protein that was targeted by darobactin (Imai et al., 2019). In addition, the use of darobactin showed an increase of periplasmic chaperones which assist in protein folding and a decrease in outer membrane proteins (OMPs) (Imai et al., 2019).
To better understand darobactin’s target and mechanism of action, mutant bacteria resistant to darobactin were grown (Imai et al., 2019). These mutants were found to have 2-3 mutations in BamA, an essential OMP (Imai et al., 2019). By mutating E. coli similarly, darobactin was unable to kill these bacteria (Imai et al., 2019). This suggests that BamA is the target of darobactin as mutations within this region were able to resist darobactin binding (Imai et al., 2019).
Knowing the mutations confer resistance to darobactin, the authors used an in vitro protein folding assay to further observe that the BAM complex was indeed directly inhibited through the action of darobactin (Imai et al., 2019). By looking into the ability of the protein to fold the protein OmpT, the authors found that darobactin inhibited this action with an IC50 of 0.68 - 1 uM, but resulted in no inhibition when the BAM complex was removed from the organism (Imai et al., 2019). Looking into the individual mutations, the authors determined the effects of three mutations with respect to the action of darobactin. The mutation M1a (G429V, T434A, G807V) resulted in a 176% increase in its IC50 and a halved folding activity (Imai et al., 2019). The mutations M2 (F394V, E435K, G443D) and M3 (T343A, Q445P, A705T) decreased folding activity to about 34% and 16% respectively where IC50 was unchanged (Imai et al., 2019). The mechanism by which mutations M2 and M3 result in darobactin resistance were not elucidated in this study (Imai et al., 2019).
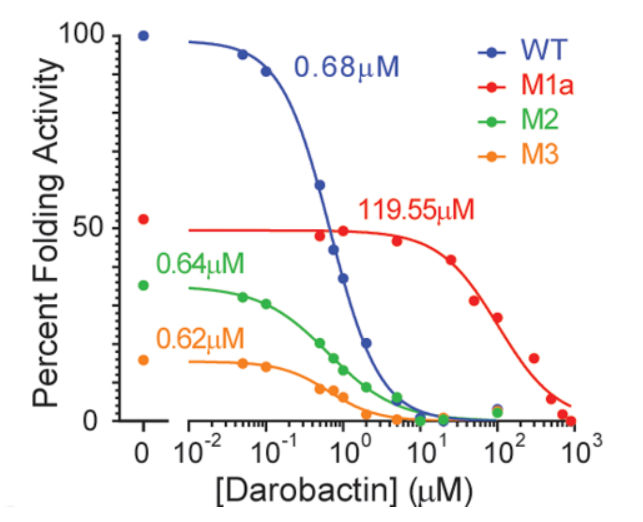
Figure 8: Three different mutations to BamA allow for resistance to darobactin with IC50 values as labeled (Imai et al., 2019).
Finally, through use of high-resolution NMR, darobactin was seen to interact with BamA such that the protein was locked into a state similar to the protein’s natural closed-gate conformation (Imai et al., 2019). The authors note that many mutations in BamA that result in resistance to darobactin occur at this gate (Imai et al., 2019). Due to this closed-gate conformation, the interaction is inferred to cause substrates to be blocked from exiting to the outer membrane (Imai et al., 2019).
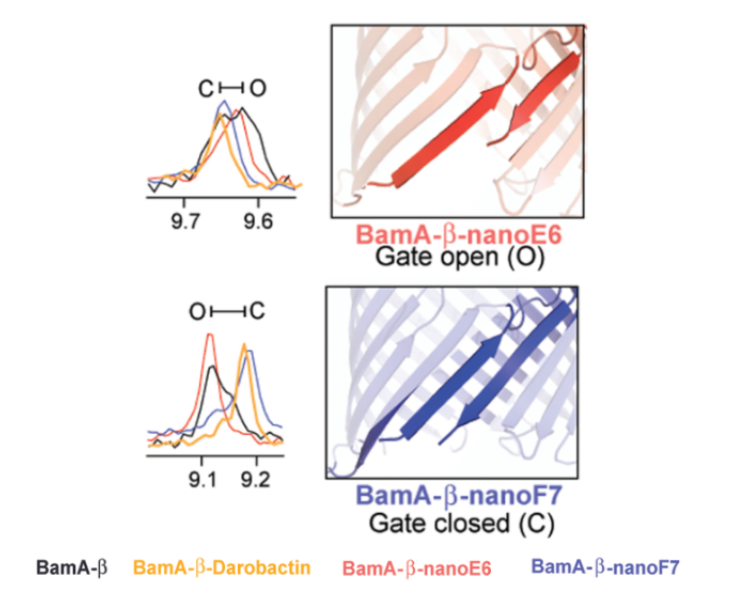
Figure 9: Using the chemical shifts in high-resolution NMR, darobactin bound to BamA shows more spectral overlap with nanoF7, a compound known to lock BamA in its closed-gate confirmation, than nano-E6, a compound known to lock BamA in its open-gate conformation (figure modified from (Imai et al., 2019)).
Animal Efficacy
The authors looked into a mouse model to check how well darobactin worked in vivo (Imai et al., 2019). Using intraperitoneal injections at a 50 mg/kg dosage, the authors found darobactin concentration in the blood peaked at 94 ug/mL where a half-life of 1 hour was observed, altogether allowing for concentrations to exist above MIC necessary to combat E. coli for an 8-hour duration (Imai et al., 2019).
A septicemia model was also used using the bacteria: WT P. aeruginosa, polymyxin-resistant P. aeruginosa (mutants species used were PAO1 and pmrB 523 C>T), WT E. coli, polymyxin-resistant E. coli (mutant species used were ATCC 25922 and Ar350 mcr-1), and K. pneumoniae that produces carbapenemase (KPC) (Imai et al., 2019).
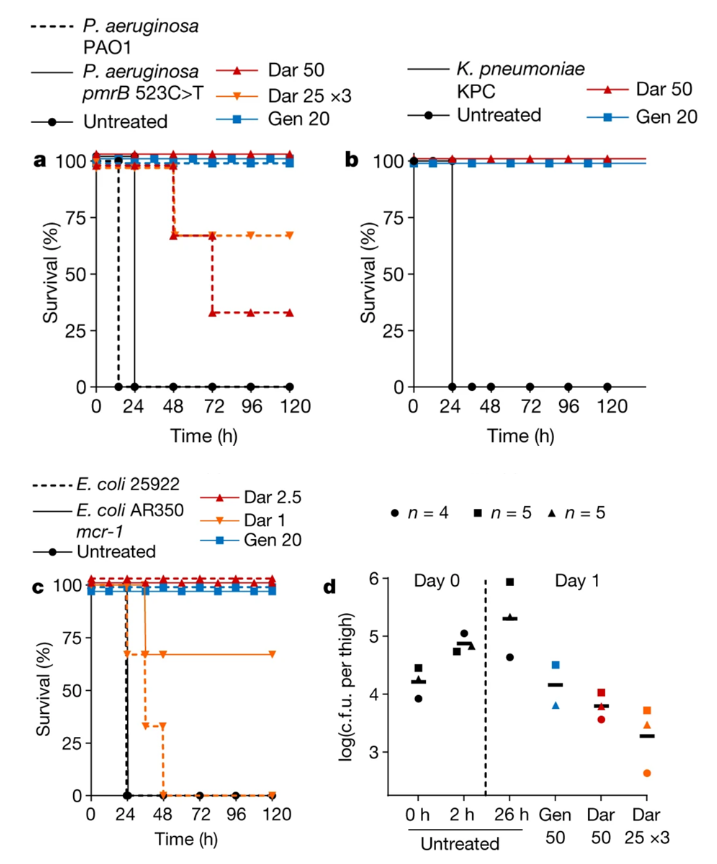
Figure 10: (a), (b), and (c): the introduction of darobactin into an infected mouse improved survival rates where the mouse would normally die within 24 hour period (Imai et al., 2019). (d) Colony forming units (c.f.u.) were measured in mouse thighs injected with darobactin revealing lower overall counts when darobactin is administered, Gentamicin was measured being the positive control (Imai et al., 2019).
Each mouse was infected with a lethal number of bacteria, where darobactin was introduced 1 hour following intraperitoneal infection (Imai et al., 2019). The authors found that the controls not given darobactin died within a 24-hour period, but the mice given the drug survived where darobactin provided complete protection against the bacteria (Imai et al., 2019). In addition, mice models were utilized to determine the virulence of darobactin resistant E. coli (Imai et al., 2019). When mice were introduced to ATCC 25922, an E. coli mutant, there was a reported 60% mortality within 24 hours (Imai et al., 2019). In comparison, E. coli BamA mutants reported no deaths within 24 hours (Imai et al., 2019). This suggests that E. coli virulence is affected by BamA mutations, be it a single or triple mutation (Imai et al., 2019). As a result, darobactin resistant bacteria could potentially cause less harm even if they are resistant due to the mutation the bacteria acquired against darobactin (Imai et al., 2019).
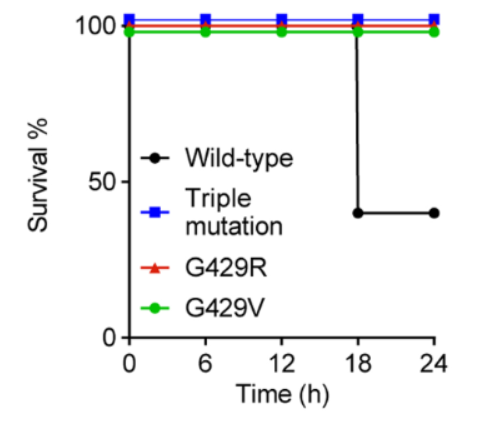
Figure 11: Survival rates of mice injected with WT E. coli, triple BamA mutant E. coli, and single BamA mutants, G429R and mutant G429V (Imai et al., 2019). Wild type E. coli has a lower survival rate of less than 50%, compared to a 100% survival rate for mutants (Imai et al., 2019). This suggests a reduction in E. coli virulence when darobactin induces a mutation to BamA (Imai et al., 2019).
Future Work
Antibiotics have greatly extended the lifespan of an average human being, ever since its introduction in the early 1900s (Center for Disease Control (CDC), 1999). Although there are currently a wide range of antibiotics, the bacteria rate of resistance has quickly surpassed the production of new antibiotics, creating an evolutionary arms race between humans and pathogenic bacteria (Lewis, 2013). However, with the identification of darobactin, there remains hope that other antibiotics can be discovered in a similar manner. By observing organisms with similar antibiotic requirements as humans, this could introduce a plethora of antibiotics that could assist with beating the race against bacterial resistance.
References
Centers for Disease Control and Prevention. (1999). Achievements in public health, 1900-1999: Control of Infectious Diseases. Centers for Disease Control and Prevention. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm4829a1.htm#fig2
Imai, Y., Meyer, K. J., Iinishi, A., Favre-Godal, Q., Green, R., Manuse, S., Caboni, M., Mori, M., Niles, S., Ghiglieri, M., Honrao, C., Ma, X., Guo, J. J., Makriyannis, A., Linares-Otoya, L., Böhringer, N., Wuisan, Z. G., Kaur, H., Wu, R., … Lewis, K. (2019). A new antibiotic selectively kills gram-negative pathogens. Nature, 576(7787), 459–464. https://doi.org/10.1038/s41586-019-1791-1
Kaur, H., Jakob, R. P., Marzinek, J. K., Green, R., Imai, Y., Bolla, J. R., Agustoni, E., Robinson, C. V., Bond, P. J., Lewis, K., Maier, T., & Hiller, S. (2021). The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Nature, 593(7857), 125–129. https://doi.org/10.1038/s41586-021-03455-w
Lewis, K. (2013). Platforms for antibiotic discovery. Nature Reviews Drug Discovery, 12(5), 371–387. https://doi.org/10.1038/nrd3975