Table of Contents
Introduction
Etiology
The Ebola virus was first identified in 1976, localized near the Ebola river, primarily in the Democratic Republic of Congo and Sudan (CDC). The virus is known to affect both humans and non-human primates, including chimpanzees, gorillas, and orangutangs. Moreover, there are 5 known strains of the Ebola virus, with the most common among humans being the Zaire virus. In addition, the Reston virus has been found to propagate disease in nonhuman primates but not among human populations (CDC).
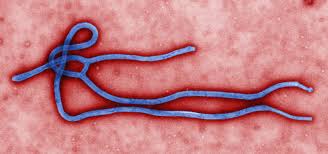
Figure 1: Zaire ebolavirus depicted as a thread-like virus under fluorescent microscopy imaging.
Ebola Strains/ Variants:
- Zaire ebolavirus
- Sudan ebolavirus
- Taï Forest ebolavirus
- Bundibugyo ebolavirus
- Reston ebolavirus
The first outbreak was recorded in 1976-1979 and was classified as Ebola Sudan, given that it occurred in Sudan. The Sudan Ebola virus primarily affected the towns of Nzara and Maridi, resulting in a 53% mortality rate (150/284). The second outbreak originated approximately 800 kilometers from Nzara in a town called Yambuku, and spread near the borders of Democratic Republic of the Congo (DRC), called the E. Zaire. The first Ebola Zaire outbreak exhibited a mortality rate of 89% and re-emerged in 1994 as the Ebola Ivory Coast strain that evolved from the Zaire strain, lasting for a period of 3 years. The next outbreak was in 1996, where the index case was a hunter who died from hemorrhagic fever. At the same time, cases of chimpanzees were reported in the surrounding forest. A second hunter was found with the virus and he transferred it to a healer and his assistant leading to the infection of several towns and villages in Gabon (DRC). The hunter contracted the virus due to ingestion of contaminated bushmeat of a gorilla carcass. Again in 2000-2004, E. Zaire and E. Sudan re-emerged and had a large impact on the mortality rate of large non-human primates including gorillas. According to statistics provided by the World Heath Organization, the largest recorded outbreak prior to the outbreak of 2014 in West Africa, was between October of 2000 to January of 2001 as a result of two simultaneous E. Sudan outbreaks. Most of the outbreaks that occurred following 2000 were due to contamination by improper handling of infected primate carcasses, leading to an epidemic chain.There is also a possibility that humans were infected by EBOV reservoir species including fruit bats, which may have led to the outbreak presented from 1976-1979 (Xavier, 2005).
Fruit bats seem to act as a natural reservoir for the Ebola virus, likely given their innate capacity to carry an antibody serum for the Ebola virus in their bodies. According to a study published by Hayman et al. in 2012, 88 non-migratory fruit bats resident to the region of Ghana, were tested for an antibody serum for the Ebola virus via PCR and ELISA assays, and 33 of them were positive for the serum and enabling them to share a commensalistic relationship with the virus. Specifically, researchers found antibodies for the virus, in addition to viral RNA fragments in three species of bats, including hammer-headed fruit bats (Kai, 2017), however, they failed to isolate the virus in culture, hence other species may play a more significant role in transmitting the virus. The virus usually spreads to primates when bat species holding the virus are eaten as bushmeat. From then on, it is easily transferred to humans as they handle dead animals or ingest them as part of their diet.
Epidemiology: Ebola Outbreak (2013-2016)
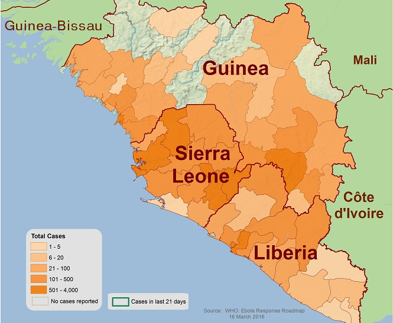
Ebola first emerged in 1976 in southern Sudan and Democratic Republic of Congo. Previous to the West Africa outbreak in 2014, there was only around 100-200 cases, while the 2014 outbreak alone had greater than 25000 cases. The West Africa outbreak involved transmission between both rural and urban areas, and across borders. The densest areas include Liberia, Guinea and Sierra Leone (Kramer et al. 2016). Differences in the magnitude of outbreaks between the 2014 outbreak and previous ones are due to external factors and not the virulence of the Ebola virus. These external factors included late diagnosis and recognition that symptoms were associated with Ebola. Another factor was the lack of experience and preparation of an Ebola outbreak in West Africa. These factors lead to the significant differences in cases (Mack et al. 2016).
Figure 2: Geographical mapping of EBOV Zaire strain localization and concentration in West Africa during the outbreak of 2014.
Symptoms and Stages of Ebola
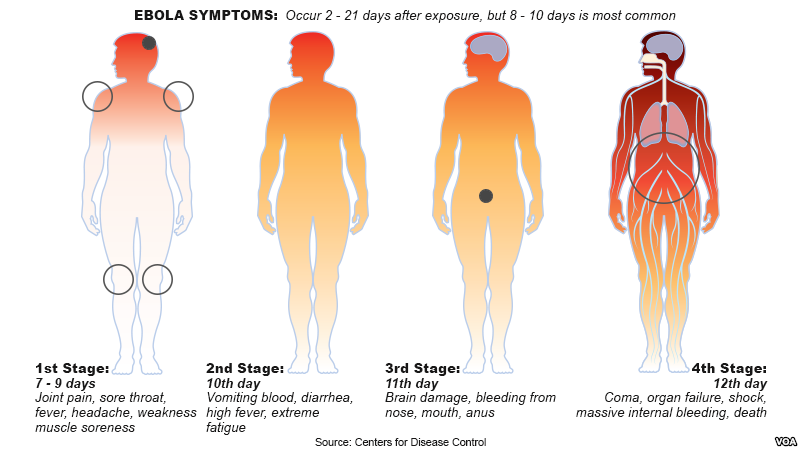
There are 4 distinguishable stages of the Ebola virus and symptoms tend to arise 2-21 days following exposure to the disease via contaminated bodily fluids in most cases. It is usual for symptoms to occur between 8-10 days after contraction of the virus.
Stage 1: Lasts from a period of 7-9 days following exposure to the virus and the incubation period. Symptoms that identify this stage include joint pain, sore throat, fever, headache, weakness, and muscle soreness.
Stage 2: Begins on approximately the 10th day of infection and is characterized by vomiting blood, diarrhea, high fever, and extreme fatigue.
Stage 3: Begins on the 11th day following transmission of the disease and is characterized by brain damage, bleeding from the nose, mouth, and anus. During this period, it is common for bodily fluids to contaminate living environments, resulting in a higher risk of transmission among humans.
Stage 4: Begins on approximately the 12th day following infection by the virus and is characterized by the occurrence of a coma, organ failure, shock, severe internal bleeding, and in cases where no immediate treatment or hospitalization is sought out, death may occur.
Figure 3: Center for Disease Control outline of stages and corresponding symptoms of Ebola following incubation period.
Overview of Symptoms According to the Centre for Disease Control and Prevention (CDC), following an asymptomatic incubation period defined as the time interval from infection to the presentation of symptoms, and ranging from 2-21 days, symptoms become apparent at approximately on average 8-10 days following initial infection. Once the initial symptoms of a fever, muscle pain, headache, sore throat, and fatigue occur, the virus is able to infect other humans and the patient is considered infectious. A comatose state may occur within one week of initial infection, in conjunction with severe hemorrhaging and external bleeding. Moreover, some symptoms including, vomiting, diarrhea, organ failure, and internal/external bleeding increase the risk of death within a short time frame given the severe loss of bodily fluids and loss of blood pressure as a direct consequence. In rare cases of survival from the virus and if treatment is sought, the virus is still able to persist in organs and ducts that contain and secrete bodily fluids, including the testes, tear ducts, and in the central nervous system. In addition, pregnant women infected with the Ebola virus are prone to transmitting the virus to their infant, specifically in the amniotic fluid, placing the infant at risk of contracting the virus (CDC)
Transmission
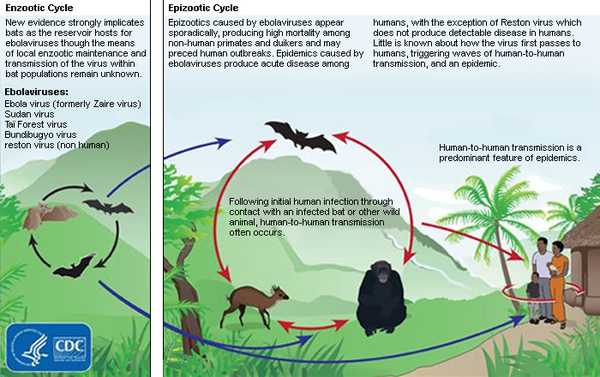
The origin of the Ebola virus is not factually known, however, researchers hypothesize that the origins of the virus lie within the species Pteropodidae, also known as fruit bats (CDC). The species likely share a relationship based on commensalism with the virus, in which the virus benefits from the nourishment that the host provides, but the host remains unaffected or harmed in any capacity. The disease is transmitted from infected fruit bats to humans, which then pass on the virus within the population. In particular, excrement from infected fruit bats often contaminates human food supplies, including fruit and meat, and if not washed or sanitized appropriately, may transmit among the human population (CDC).
Transmission among humans is often through direct contact with infectious bodily fluids and secretions, including blood, semen, excrement, saliva, and breastmilk through broken skin or mucous membranes of the eyes, nose, and mouth. Specific examples of transmission among humans include:
- Blood, urine, saliva, sweat, feces, vomit, breast milk, and semen of an infected individual or a patient that has died from Ebola.
- Contaminated needles or syringes that contain infectious bodily fluid residues and exposure of these fluids to exposed skin or mucous membranes may cause transmission among humans.
- Engagement in sexual intercourse with a man who has recovered from symptoms of Ebola, in particular, contact with their infectious semen (CDC).
Figure 4: Centre of Disease Control and Prevention guide to Ebola Virus transmission from reservior hosts (fruit bats) to secondary hosts that can pass virus in epidemic stages.
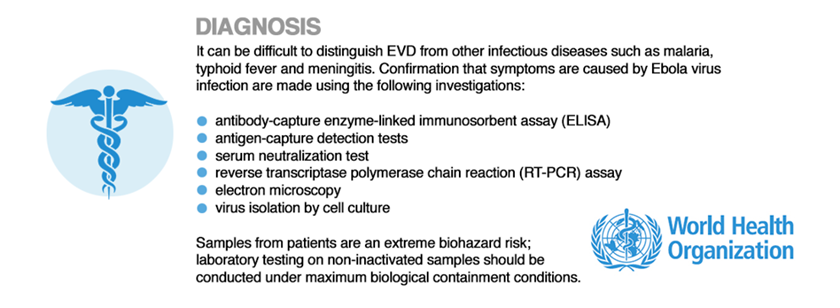
Diagnosis
Effective diagnosis of the Ebola virus in the region of sub-Saharan Africa/ Western Africa is often difficult given the lack of infrastructure and disease screening technologies and resources. Regardless, in general, diagnostic blood tests and laboratory assays can be conducted to assess the patient for Ebola, which may be useful during the asymptomatic period to offer preemptive treatment protocol and to distinguish the symptoms of the Ebola virus from the symptoms caused by malaria, meningitis, and typhoid (WHO 2014). The World Health Organization has implemented a structured guideline for laboratory testing involving techniques such as:
- Blood testing
- Antibody-capture enzyme-linked immunosorbent assay (ELISA)
- Antigen-capture detection tests
- Serum neutralization test
- Reverse transcriptase polymerase chain reaction (RT-PCR) assay
- Electron microscopy
- virus isolation by cell culture
These tests may aid in the detection of abnormalities in the blood plasma or immune response, and facilitate the detection of viral antigens and proteins (EBOV glycoproteins) characteristic of the Ebola virus (Broadhurst et al., 2016). For instance, initial blood tests may reveal an abnormal leukocyte count. This is due to a reduction in viral antigen-presenting dendritic cells, and consequently resulting in low levels of T immune cells and B immune cells that respond to dendritic cells in the blood plasma. Essentially, the virus is effectively able to target, infect, and destroy dendritic cells, which results in reduced activation of the T and B immune cells that respond to the protein profiles presented on dendritic cells. As a result, the immune system of a patient infected with the Ebola virus is often compromised and co-morbidities or supplemental infection during infection with Ebola can accelerate morbidity and mortality (Bray & Geisbert, 2005). Furthermore, the Ebola virus may target hepatocytes, resulting in the detection of low levels of blood coagulation proteins and enzymes by these protein screening tests (Falasca et al., 2015).
A new diagnostic test called the ReEBOV Antigen Rapid Test Kit (Corgenix, USA) was studied by the World Health Organization under their Emergency Assessment and Use, to determine performance levels and gauge safety when implemented in the field. The test was developed to assist in rapid diagnostic testing amidst the outbreak in 2014 and to avoid the expense and time-dependence associated with lab diagnostic screening. The ReEBOV kit can detect viral surface glycoproteins and determine positive results within 15 minutes with an accuracy of 92% as opposed to laboratory screening tests that detect viral DNA/RNA and take 12-24 hours to provide results. Though this novel testing method is less accurate than most laboratory screening tests, it is efficient and rapid, requiring no source of electricity and can be used in regions with poor healthcare infrastructure or in remote regions. Results of the ReEBOV test should be supplemented by advanced laboratory tests or blood tests where accessible (Broadhurst et al., 2015).
Figure 5: World Health Organization guide to effective diagnostic laboratory testing of the Ebola virus.
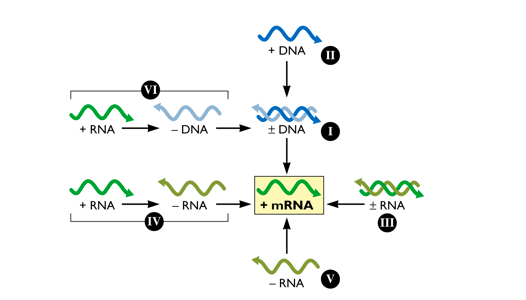
Baltimore Classification of Viruses
Viruses can either be made up of RNA or DNA. In order to be translated by host proteins however the viral genome must be converted into a positive strand mRNA template. The Baltimore classification is made up of 5 classes of viruses and is based on how the viral genome transforms itself into the positive strand mRNA template (Baltimore, 1971). Class I: are composed of viruses with double stranded DNA genomes. These viruses insert their genomes into the nucleus and are highly dependent on host machinery such as DNA dependent DNA polymerase (DdDp) and RNA dependent RNA polymerase (RdRp) for replication and transcription of the positive mRNA strand respectively (Dimmock, 2007). Class II: are composed of viruses with single stranded DNA genomes. Once inside the cell, either through virus or host machinery a double stranded DNA intermediate will be produced. This double stranded DNA intermediate will then be able to serve as a template for replication, which will construct a single stranded genome and transcription, which will produce a positive mRNA strand that can be translated (Dimmock, 2007). Class III: are composed of viruses with segmented double stranded RNA genomes. There are usually 11-12 segments that are transcribed to produce monocistronic mRNAs- each template codes for a separate protein. The virus uses host RdRp to replicate the segmented genome (Dimmock, 2007). Class IV: are composed of viruses with segmented single stranded positive stranded RNA genomes. The mRNA genome can be replicated by host RdRp and can be directly transcribed by host machinery since it is already in positive sense formation. The genome is a polycistronic mRNA template which codes for one large protein that is then cleaved into individual components (Dimmock, 2007). Class V: are composed of viruses with single stranded negative stranded RNA genomes. There are two main subclasses, segmented and non-segmented. The non-segmented mRNA genomes are transcribed into several monocistronic templates to form individual viral proteins. The genome is also transcribed into a positive stranded mRNA template via viral RdRp which can be then used to produce genome copies for viral progeny (Dimmock, 2007).
Figure 6: Baltimore classification is based upon how the viral genome is transformed into a positive stranded mRNA template to allow for vial protein translation.
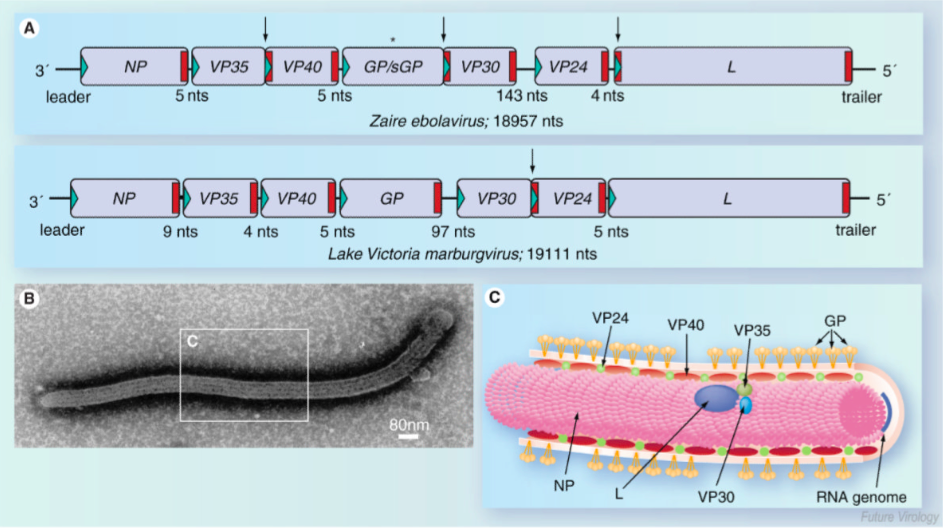
Virion Structure
Ebola virus (EBOV) is a filamentous enveloped virus containing a negative strand RNA genome 19 kb in length (Falasca et al., 2015). It is transcribed into seven monocistronic mRNA templates that encodes for a nucleoprotein (NP), glycoprotein (GP), RNA-dependent RNA polymerase (L), and four structural proteins termed VP24, VP30, VP35, and VP40(Falasca et al., 2015). The genome is arranged in such a way that the genes located near the 3' end will be transcribed at the highest rate and the genes at 5' end are transcribed at the lowest rate. Therefore regarding to Figure _A, NP gene will be transcribed the most and the L gene the least (Muhlberger, 2007). The surface GP has important roles in virus infection and pathogenesis, as it is responsible for binding and fusion to host cells. The RNA-dependent-RNA-polymerase (L) is responsible for replicating viral RNA. NP embeds the viral genetic material and and along with proteins VP30, VP35, and VP24 forms a fully functional nucleocapsid. VP40 is required viral cellular egress and VP24 and VP35 also play a key role in suppressing host immunity (Falasca et al., 2015).
Replication: As perilously discussed, in order for replication to occur, a positive stranded mRNA template must be constructed. The EBOV virus encodes its own RpDp to produces this positive stranded mRNA template. Replication occurs in the host cytoplasm and is dependent on three nucleocapsid proteins; NP, VP35 and L. Referring to Figure _C, VP35 acts as a linker protein and mediates the interaction between N and L proteins. VP35 guides L (main catalytic component) to the encapsidated RNA genome to allow for replication (Mulberger, 2007).
Transcription: Transcription is also occurs in the host cytoplasm and is mediated by NP, L and Phosphoprotein (P) VP30. VP30 has dual function- it regulates nucleocapsid assembly by binding to NP as well as functions as a transcription activator via its zinc-binding domain. In vitro, loss of function of the zinc-binding domains led to a loss of transcription activation. Though the exact function of VP30 is unknown, it hypothesized to be involved in transcription initiation or early anti-termination. In order to be translated by host machinery, the virus mRNA template must have a 5' cap. It is inefficient to encode for components viruses can attain from the host, so they don't encode a 5' cap in their genome. Instead, an endonuclease cleaves 5' caps off of host mRNA and primes it to the viral mRNA species. The second component needed to ensure translation is the addition of a poly-A-tail. The termination codon is followed by a stretch of uridine residues that codes for the poly-A-tail and causes the polymerase to “fall off” the mRNA template (Mulberger, 2007).
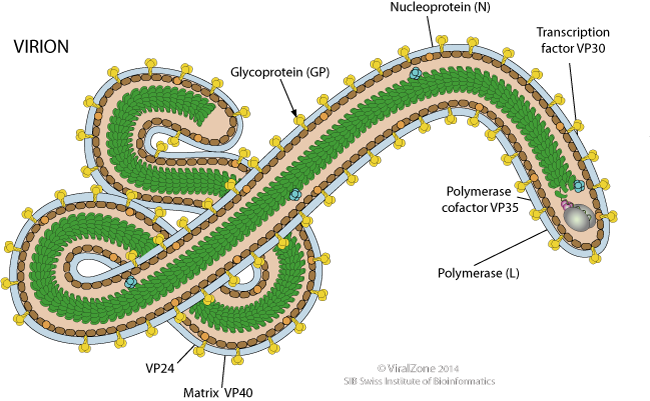
Figure 7: Depiction of the structural features of the Ebola virus, including surface glycoprotein, nucelocapsid, and RNA-dependent RNA polymerase.
Figure 8: Genes on monocistronic RNA genome of Ebola virus coding for key components of viral nucleocapsid (coded by NP gene on 3' end), glycoprotein epitope for viral endocytosis (coded by GP gene), and RNA-dependent RNA polymerase (coded by L gene).
Pathophysiology of Ebola
Macrophages and dendritic cells are early targets of EBOV. Extensive infection of these cells leads to overproduction of inflammatory mediators and chemokines such as IL-1, IL-6, IL-8, TNF-a, and monocyte chemotactic protein-1 (MCP-1)(Marcinkiewicz et al., 2014). This “cytokine storm” is responsible for attracting neutrophils and eosinophils to the infected tissues, which subsequently induces coagulopathy (impaired coagulation) and increases endothelial permeability (Marcinkiewicz et al., 2014). Additionally, the infection of antigen presenting cells leads to extensive apoptosis of T cells and natural killers cells. Another key point of infection is the suppression of the type I interferon (IFN) response, via the non structural proteins, VP24 and VP35. Type I IFNs are responsible for inducing transcription of a large group of genes which play a major role in host resistance to viral infection and activate key components of the innate and adaptive immune systems including; antigen presentation and the production of cytokines that activate T cells, B cells, and natural killer cells. Furthermore, VP35 inhibits maturation of dendritic cells by interfering with the RIG-I signaling pathway to prevent upregulation of MHC I and MHC II and the costimulatory molecules CD40, CD80, and CD86, thus impairing antigen presentation to CD8+ and CD4+ T cells and T-cell activation and thereby impeding linkage of the innate and adaptive immune responses (Ansari, 2014). The evasion of the immune system allows the virus to replicate unhindered and therefore permits subsequent dissemination of viral progeny. Infected antigen presenting cells (macrophages and dendritic cells) travel to the lymph nodes and to the spleen, where further replication takes place and dissemination to the rest of the body is made possible via the blood stream (Ansari, 2014). Viremia allows the virus to infect the liver, the adrenal gland and the gastro-intestinal tract (leading to diarrhea). Infection of the liver leads to the dysregulation of coagulation proteins and infection of the adrenal gland compromises the ability to make blood pressure regulating steroids, in tandem this dissemination leads to hemorrhagic fever, multi-organ failure, terminal shock and death (Ansari, 2014).
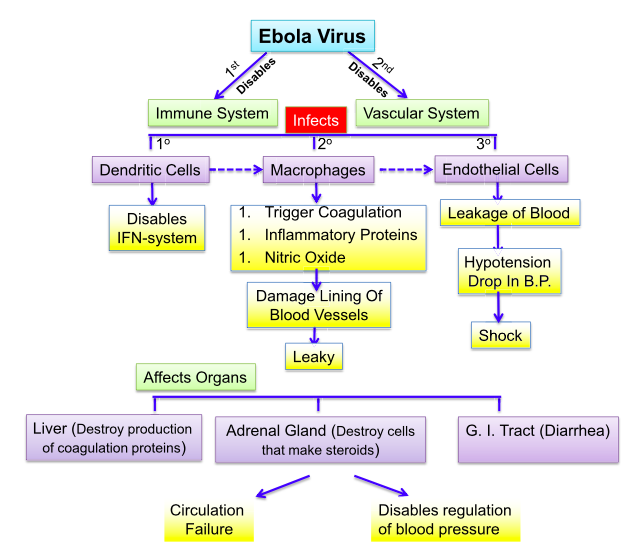
Figure 9: The cascade of pathological events that results in the rapid severity of Ebolavirus infection.
Treatments
Supportive Care
To date, no EBOV-specific therapy has achieved regulatory approval for use in humans (Baseler et al., 2017). Consequently, supportive care remains the mainstay of treatment. Optimal intensive supportive therapy for EBOV infection requires near-continuous bedside care to closely monitor vital signs, fluid and electrolyte status, and clinical complications. Noninvasive monitoring of blood pressure, cardiac telemetry, and oxygen saturation, greatly facilitate care. Pharmacotherapy includes the use of antiemetics, antidiarrheal medication, analgesics, sedatives, antibiotics, and antimalarial medications. Nutrition may be provided via enteral or parenteral routes (Baseler et al., 2017).
ZMapp
Developed by Mapp Biopharmaceutical, Inc., based in San Diego, is composed of three different laboratory-made proteins called monoclonal antibodies. The treatment is designed to prevent the progression of EVD within the body by targeting the main surface glycoprotein of the Ebola virus that is essential in facilitating viral endocytosis of the host cell. The monoclonal antibodies work to bind complementarily to the viral glycoprotein epitope and inhibit binding to host cell receptors, thus preventing endocytosis and viral infection of the host cell. Earlier studies in nonhuman primates demonstrated that ZMapp had strong antiviral activity and prevented death when administered as late as five days after experimental infection with Zaire ebolavirus. It is tested on animals and results indicate it helps alleviate the fever, viraemia and abnormalities of blood count. It also reverses elevated liver enzymes and mucosal haemorrhages (Marcinkiewicz et al., 2014).
rVSV-ZEBOV
NewLink Genetics of Ames from Iowa collaborating with Canadian National Institute of Public Health genetically engineered the vesicular stomatitis virus (VSV), that has a natural tropism for livestock (Marcinkiewicz et al., 2014). VSV was attenuated, engineered to be replication-competent and to express the glycoprotein from a Zaire strain of Ebola virus (ZEBOV) in replacement of its own glycoprotein (rVSV-ZEBOV). Vaccination induces replication of these genetically engineered viral particles (rVSV-ZEBOV)(Regules et al., 2017). The ZEBOV glycoprotein is responsible for binding and fusion to host target cells, and is the viral epitope that is targeted by host cytotoxic T-cells and neutralizing antibodies. Therefore, exposing patients to the ZEBOV glycoprotein in the form of an attenuated and genetically engineered vesicular stomatitis virus, allows for the activation of adaptive immunity against ZEBOV without the risk of infection (Regules et al., 2017). In fact, the rVSV-ZEBOV vaccine has been shown to be attenuated in normal and immunocompromised nonhuman primates in safety and immunogenicity studies. In addition, multiple studies in cynomolgus macaques have shown that a single administration of the vaccine confers a high level of protection. There are various methods of vaccine delivery (oral, intranasal, or intramuscular) and all variants have shown protective efficacy in animal models (Regules et al., 2017).
Persistence of Ebola
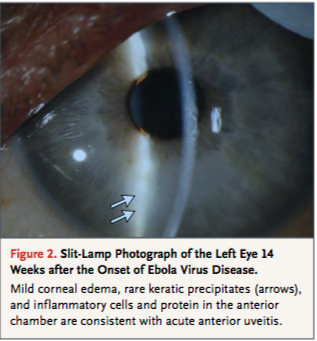
Though the subject of Ebola persistence has not been thoroughly researched, there have been cases where patients experience post-infection symptoms and have had traces of EBOV in sperm and ocular aqueous humour. In 2015 a 43 year old physician who was helping with the Ebola breakout in Sierre Leone contracted the disease. He was transferred to the Emory University Hospital in Atlanta and treated with an interfering RNA antiviral agent. His case, similar to many, also had the presence of post-infection symptoms. His case however was intensely monitored to accumulate knowledge on how to increase survival rates and prevent the spreading of EBOV. After almost a month of being hospitalized, the patient was discharged after regaining mental status and function of kidneys. His urine and blood samples came up negative for EBOV but his sperm showed traces of the virus. The patient was then advised abstain from unprotected sex for at least 3 months. Post-discharge he acquired new symptoms such as lower back pain and ophthalmic symptoms (ocular burning, foreign-body sensation and re-adjustment of his glasses prescription). His symptoms worsened over time with his visual acuity worsening to 20/400 and being diagnosed with vitrifies and anterior uveitis. His ocular pressure became significantly elevated to 44 mm Hg (normal range between 10-21 mm Hg). To control for this, a paracentesis of the anterior chamber was performed. The specimen was then analyzed and tested positive for EBOV. Fortunately there were no left over traces of EBOV found in the conjunctiva and tear ducts so he was not contagious. He was then treated with ophthalmic drops to further lower pressure. Within 3 months, all symptoms cleared and the patient's visual acuity was recovered. This particular case provides insight on how to treat post-infection symptoms for Ebola patients as well as reassures the notion that once the EBOV virus has passed from the blood and urine, the patient is no longer contagious (Varkey, 2015).
Figure 10: Ebola virus can persist in organs such as eye, namely in the vitreous or aqueous humour, leading to high ophthalmic pressure, uveitis, and diminished visual accuity.
Lesson Learned From the Ebola Outbreak
The Ebola outbreak has demonstrated the lack of preparation the world has for future epidemics. This is especially concerning since the epidemic could involve a virus the spreads more easily than Ebola, such as the mode of air borne transmission. Ebola has shown that there is a need for better communication between nations to control spread and to coordinate activities. This coordination between nations will allow rapid action and decision making on a global level for the next epidemic. Another step needed to prepare for the next epidemic is to allocate more funds to research that develops new tools for diagnosis and treatment of viruses. Furthermore, there needs to be an improvement in health care facilities in third and second world countries to control the spread and to be better equipped to deal with the virus. All these steps involve the cooperation of many nations to control an epidemic (Gates 2015).
Other ways to further prevent another viral outbreak includes minimization of unsafe handling of bush meat, to decrease spread of zoonotic diseases into human populations. Additionally, promotion of biosafety by providing health care workers with education on infection control, and increased supplies. There can also be a reduction in transmission by practicing safe burial methods, and educating the community about the disease. The transmission can be also reduced by creating adequate isolation and treatment centres, and provide health care workies with efficient protective materials and essential supplies. Furthermore, there needs to be an improved method for quick diagnosis and transport for samples. Lastly, a data system in which disease cases and information can be reported for disease surveillance on a global level (Kaushik et al. 2016).
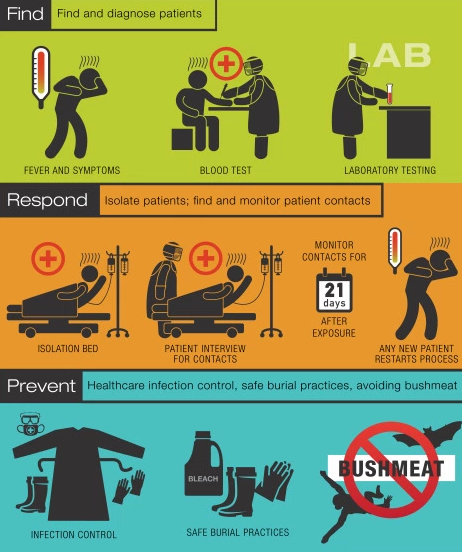
Figure 11: Guide to preventative and disease controlling measures to manage or prevent viral epidemics.
Presentation
References
Ansari, A. A. (2014). Clinical features and pathobiology of Ebolavirus infection. Journal of Autoimmunity, 55, 1-9. http://dx.doi.org/10.1016/j.jaut.2014.09.001
Biek R, Walsh PD, Leroy EM, Real LA. Recent Common Ancestry of Ebola Zaire Virus Found in a Bat Reservoir. PLOS Pathogens. [accessed 2017 Nov 24]. http://journals.plos.org/plospathogens/article?id=10.1371%2Fjournal.ppat.0020090
Baseler, L., Chertow, D. S., Johnson, K. M., Feldmann, H., & Morens, M. (2017). The Pathogeneis of Ebola Virus Disease. Annual Review of Pathology, 12, 387-418. https://doi.org/10.1146/annurev-pathol-052016-100506
Bray, M., Geisbert, TW. (2005). Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. The International Journal of Biochemistry and Cell Biology, 7(8):1560-1566. https://www.ncbi.nlm.nih.gov/pubmed/15896665
Broadhurst, MJ., Kelly, JD., Miller, A., Semper, A., Bailey, D., Groppelli, E., Simpson, A., Brooks, T., Hula, S., Nyoni, W., Sankoh, AB., Kanu, S., Jalloh, A., Ton, Q., Sarchet, N., George, P., Perkins, MD., Wonderly, B., Murray, M., Pollock, NR. (2015). ReEBOV Antigen Rapid Test kit for point-of-care and laboratory-based testing for Ebola virus disease: a field validation study. The Lancet, 386(9996): 867-874. http://www.thelancet.com/journals/lancet/article/PIIS0140-6736(15)61042-X/abstract
Broadhurst, MJ., Brooks, TJ., Pollock, NR. (2016). Diagnosis of Ebola virus disease: past, present, and future. Clinical Microbiology Reviews, 29(4):773-793. http://cmr.asm.org/content/29/4/773.full
Centers for Disease Control and Prevention (CDC). Ebola (Ebola Virus Disease). CDC. [accessed 2017 Nov 18]. https://www.cdc.gov/vhf/ebola/about.html
Dimmock, N. (2007). Introduction to Modern Virology. Victoria, Australia. Blackwell Publishing.
Falasca, L., Agrati, C., Petrosillo, N., Di Caro, A., Capobianchi, M. R., Ippolito, G., & Piacentini, M. (2015). Molecular mechanisms of Ebola virus pathogenesis: focus on cell death. Cell Death and Differentiation, 22(8), 1250–1259. http://doi.org/10.1038/cdd.2015.67
Gates, B. The next epidemic—lessons from Ebola. N Engl J Med. 2015; 372: 1381–1384
Hayman, D. T. S., Yu, M., Crameri, G., Wang, L.-F., Suu-Ire, R., Wood, J. L. N., & Cunningham, A. A. (2012). Ebola Virus Antibodies in Fruit Bats, Ghana, West Africa. Emerging Infectious Diseases, 18(7), 1207–1209. http://doi.org/10.3201/eid1807.111654
Kai KupferschmidtJun. 1, 2017 , 1:00 PM, 20 2017 JKN, 20 2017 KKN, 20 2017 ESN, Marc Heller, E&E NewsNov. 20, 2017, 17 2017 MWN, 20 2017 N, 17 2017 N, 16 2017 N. Hunting for Ebola among the bats of the Congo. Science | AAAS. 2017 Jul 26 [accessed 2017 Nov 24]. http://www.sciencemag.org/news/2017/06/hunting-ebola-among-bats-congo
Kaushik, A., Tiwari, S., Jayant, R. D., Marty, A., & Nair, M. (2016). Towards Detection and Diagnosis of Ebola Virus Disease at Point-of-Care. Biosensors & Bioelectronics, 75, 254–272. http://doi.org/10.1016/j.bios.2015.08.040 Marcinkiewicz, J., Bryniarski, K., & Katarzyna, N. (2014). Ebola Haemorrhagic Fever Virus: Pathogenesis, Immune Responses, Potential Prevention. Folia Medica Cracoviensia, 3, 39-48. Retrieved from http://www.fmc.cm-uj.krakow.pl/pdf/54_3_39.pdf
Kramer, A. M., Pulliam, J. T., Alexander, L. W., Park, A. W., Rohani, P., & Drake, J. M. (2016). Spatial spread of the West Africa Ebola epidemic. Royal Society Open Science, 3(8), 160294. http://doi.org/10.1098/rsos.160294
Mack, A., Snair, M. R., & Mundaca-Shah, C. (2016). The ebola epidemic in West Africa: proceedings of a workshop. Washington, DC: The National Academies Press.
Muhlberger, E. (2007). Filovirus replication and transcription. Retrieved from https://www.futuremedicine.com/doi/abs/10.2217/17460794.2.2.205.
Regules, J., Beigel, J., Paolino, M., Voel, J., Castellano, A., Hu, Z.,…Thomas, J. (2017). A Recombinant Vesicular Stomatitis Virus Ebola Vaccine. The New England Journal of Medicine, 376, 330-341. Retrieved from http://www.nejm.org/doi/full/10.1056/NEJMoa1414216af=R&rss=currentIssue#t=article
Varkey, J. B., Shantha, J. G., Crozier, I., Kraft, C. S., Lyon, G. M., Mehta, A. K., … & Ströher, U. (2015). Persistence of Ebola virus in ocular fluid during convalescence. New England Journal of Medicine, 372(25), 2423-2427. Retrieved from http://www.nejm.org/doi/full/10.1056/NEJMoa1500306#t=article.
World Health Organization (WHO). (2014). Laboratory diagnosis of Ebola virus disease. [accessed 2017 Nov 18]. http://www.who.int/csr/resources/publications/ebola/laboratory-guidance/en/
Xavier Pourrut, Brice Kumulungui, Tatiana Wittmann, Ghislain Moussavou, André Délicat, Philippe Yaba, Dieudonné Nkoghe, Jean-Paul Gonzalez, Eric Maurice Leroy, The natural history of Ebola virus in Africa, In Microbes and Infection, Volume 7, Issues 7–8, 2005, Pages 1005-1014, ISSN 1286-4579, https://doi.org/10.1016/j.micinf.2005.04.006. (http://www.sciencedirect.com/science/article/pii/S1286457905001437)